How to Recognize Labeling Errors in Medical Data and Request Corrections
 Jan, 11 2026
Jan, 11 2026
When you're working with medical data - whether it's patient records, diagnostic images, or clinical notes - the labels attached to that data can make or break an AI model. A single mislabeled X-ray or incorrectly tagged symptom can lead to a system missing a tumor, misdiagnosing a condition, or recommending the wrong treatment. Labeling errors aren't just typos. They’re systemic problems that quietly degrade performance, often going unnoticed until a model fails in production. The good news? You don’t need to be a data scientist to spot them. You just need to know what to look for and how to ask for corrections the right way.
What Labeling Errors Actually Look Like in Medical Data
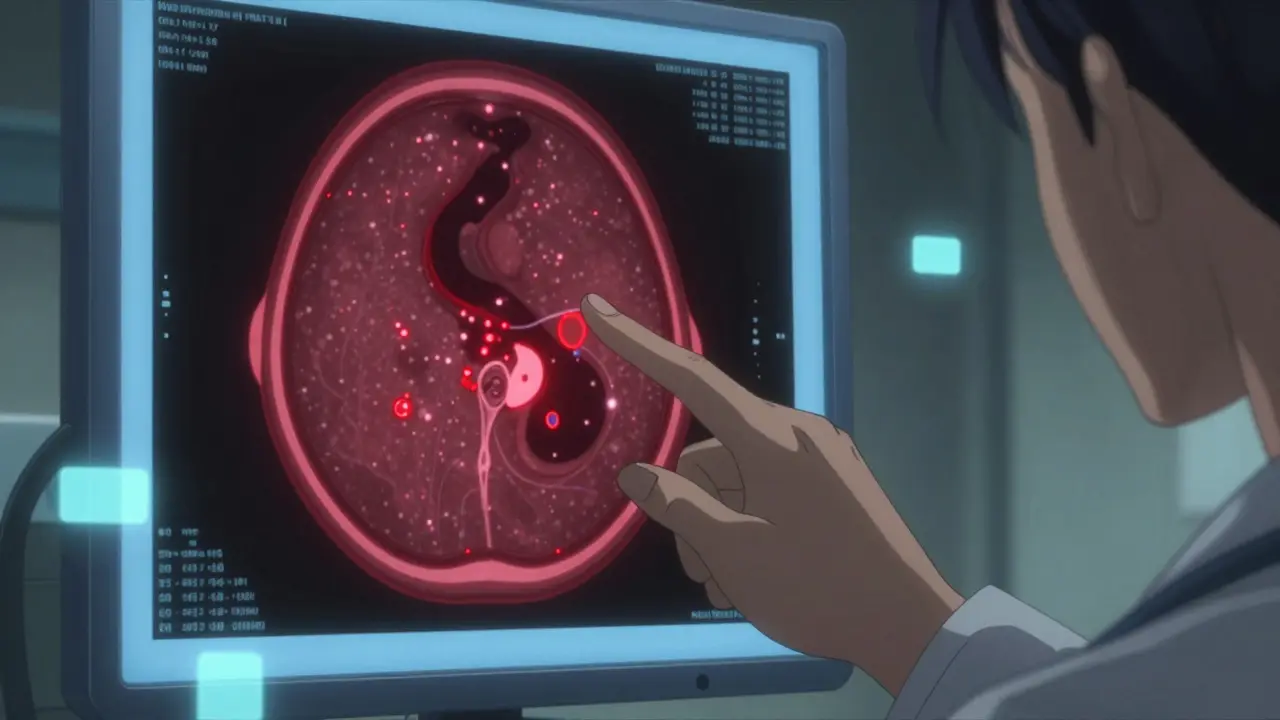
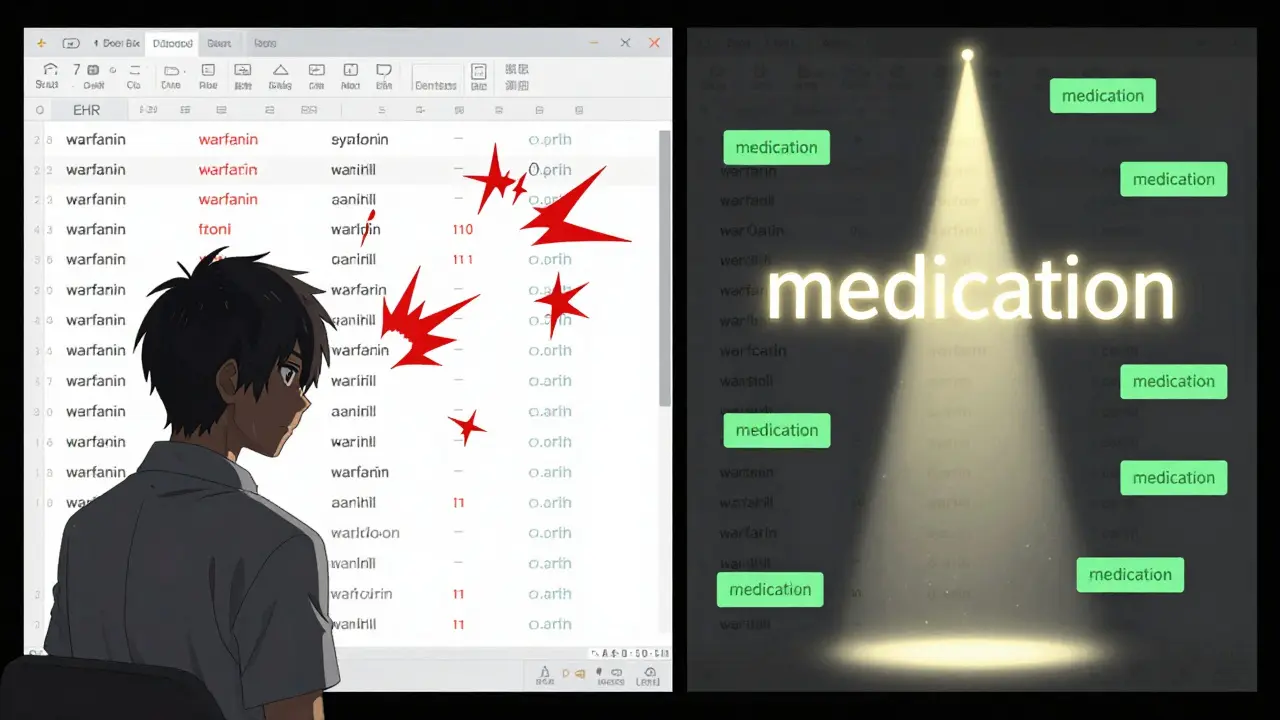
Labeling errors in medical datasets aren’t random. They follow patterns. In a 2024 study of 12,000 annotated medical images from Australian hospitals, researchers found that 41% of errors involved incorrect boundaries - meaning a tumor was labeled as smaller or larger than it actually was. Another 33% were outright misclassifications: a benign lesion marked as malignant, or pneumonia labeled as a normal chest X-ray. And 26% of cases? The abnormality was simply missing from the annotation entirely. In text data, like discharge summaries or doctor’s notes, errors show up differently. Entity recognition systems often fail because a patient’s medication isn’t tagged at all, or a drug name is labeled as a condition. For example, "aspirin" might be tagged as a symptom instead of a medication. Or worse - a drug like warfarin gets labeled as "anticoagulant," which is correct, but the system then ignores all other anticoagulants because the training data never included them. These aren’t just "mistakes." They’re data quality failures with real-world consequences. A 2023 analysis from the Royal Melbourne Hospital showed that models trained on datasets with uncorrected labeling errors had a 22% higher false-negative rate in detecting early-stage sepsis. That’s not a bug. That’s a safety risk.How to Spot These Errors Yourself
You don’t need fancy tools to catch errors. Start with the basics:- Check for missing annotations. Look at a sample of 20-50 records. Are there obvious abnormalities that weren’t labeled? If a patient has a documented skin rash but no annotation in the image or text, that’s a red flag.
- Look for inconsistent tagging. Is "hypertension" sometimes labeled as "high blood pressure"? Are different annotators using different terms for the same thing? Inconsistency breeds confusion in models.
- Watch for extreme class imbalance. If 95% of your labeled cases are "normal" and only 5% are "abnormal," the model will learn to ignore the rare cases. That’s not a labeling error per se - but it’s often caused by annotators avoiding rare cases because they’re hard to identify.
- Compare labels with source documents. If you’re working with clinical notes, pull up the original EHR entry. Does the label match what was written? If the note says "patient reports chest pain for 3 days," but the label says "acute MI," that’s a mismatch. The annotation added information not in the source.
- Spot ambiguous cases. Is there a case where two labels could both make sense? For example, a lung nodule that’s too small to confidently call benign or malignant? If annotators are unsure, the model will be too.
One real example from a Sydney-based diagnostic lab: An AI model was trained to detect diabetic retinopathy from retinal scans. The annotators were told to label "any visible microaneurysms." But some annotators only labeled large ones, while others flagged tiny dots. The model ended up over-predicting microaneurysms because it learned to see them everywhere. The fix? A new guideline with clear examples - and a re-annotation of the first 1,000 images.
How to Ask for Corrections Without Being Ignored
Most labeling teams are overwhelmed. If you just say, "This is wrong," you’ll get ignored. You need to make it easy for them to fix it.- Be specific. Don’t say: "The labels are bad." Say: "In image #4823, the left lung nodule is labeled as size 4mm, but the scale bar shows it’s actually 8mm. The annotation box is too small by 50%.
- Provide context. Attach the original clinical note or radiology report. Say: "This matches the radiologist’s report dated 12/15/2025, which states: '2.1 cm solid nodule in right upper lobe.'"
- Use a template. Create a simple form:
- Dataset ID:
- Annotation ID:
- Expected label:
- Current label:
- Evidence: [attach screenshot or document excerpt]
- Why this matters: [e.g., "This could cause false negatives in early cancer detection"]
- Don’t blame. Say: "I noticed this might be an inconsistency. Could we review it?" Not: "You labeled this wrong."
At the Royal Adelaide Hospital, a clinical informatics team started using this template. Within six weeks, their labeling error rate dropped from 11.4% to 3.1%. Why? Because the annotators felt supported, not attacked.
Tools That Help - and Which Ones to Use
You don’t have to do this manually every time. Tools exist to flag errors before they become problems.- cleanlab - This open-source tool uses statistical methods to find likely mislabeled examples. It works best with classification tasks like diagnosing pneumonia or classifying drug reactions. It doesn’t tell you what the correct label is - just which ones are probably wrong. You still need a human to confirm.
- Argilla - A web-based platform that lets you review flagged errors directly in your browser. It integrates with cleanlab and lets you correct labels in place. Ideal for teams without coding skills.
- Datasaur - Built for annotation teams. It has a built-in error detection feature that flags inconsistencies as you go. Great if you’re labeling text data like discharge summaries or patient surveys.
But here’s the catch: none of these tools work unless you feed them clean data. If your training set is full of errors, the tool will just flag more errors - and you’ll drown in noise. Start small. Pick one dataset. Fix the top 10% of errors. Then retrain. You’ll see results faster than you think.

Why This Matters More Than You Think
The FDA now requires that any AI system used in medical diagnosis must include documentation of data quality controls - including label error detection and correction. That’s not a suggestion. It’s a regulation. In Australia, the Therapeutic Goods Administration (TGA) is moving in the same direction. If your organization is building or using AI for clinical decision-making, you’re already under pressure to prove your data is reliable. But beyond compliance, there’s ethics. A mislabeled label can mean a patient gets the wrong treatment. Or no treatment at all. The cost of a labeling error isn’t just a lower accuracy score. It’s a broken trust.What to Do Next
Start today:- Pick one dataset you’re working with - even if it’s small.
- Review 20 random samples manually. Look for the five error types above.
- Write up three clear correction requests using the template.
- Share them with your annotation team. Don’t ask for a full re-annotation. Just ask: "Can we fix these three?"
- Track the change in model performance after the fix.
Most teams don’t realize how much they’re losing to sloppy labels. You don’t need to fix everything. Just fix the ones that matter. One corrected label can mean one patient gets the right care.
How common are labeling errors in medical datasets?
Labeling errors are extremely common. Studies show medical datasets have error rates between 8% and 15%, with some areas like radiology and pathology reaching up to 20%. A 2024 analysis of 12,000 annotated medical images found that 41% of errors involved incorrect boundaries, 33% were misclassified labels, and 26% were completely missing annotations.
Can AI tools automatically fix labeling errors?
AI tools like cleanlab can flag likely errors with 75-90% accuracy, but they cannot automatically fix them. They highlight examples that are statistically unlikely to be correct - but only a human with domain knowledge can determine the right label. For example, a tool might flag a lung nodule as mislabeled, but only a radiologist can say whether it’s benign, malignant, or a scanning artifact.
What’s the biggest mistake people make when correcting labels?
The biggest mistake is assuming one round of corrections is enough. Labeling errors often stem from unclear instructions, not human error. If you fix the labels but don’t update the guidelines, the same mistakes will reappear. Always pair corrections with updated annotation manuals and training for annotators.
Do I need to retrain the whole model after fixing labels?
Not always. If you’ve only corrected a small number of labels (under 5% of the dataset), you can often fine-tune the model with just the corrected examples. But if you’ve fixed more than 10% - especially if they were high-confidence cases - you should retrain from scratch. Partial training can introduce new biases.
Why does label quality matter more than model complexity?
MIT’s Data-Centric AI research showed that correcting just 5% of label errors in a medical imaging dataset improved diagnostic accuracy more than upgrading from a ResNet-50 to a ResNet-101 model. No matter how advanced your AI is, garbage data leads to garbage results. Clean labels are the foundation - not an afterthought.


Jennifer Phelps
January 12, 2026 AT 09:59Just reviewed 30 chest X-rays from our hospital's dataset and found 7 with mislabeled nodules. One had a 12mm nodule labeled as 4mm. The scale bar was right there in the corner. How is this still happening?
These aren't typos. They're systemic failures. We're putting patients at risk and no one seems to care.
Sona Chandra
January 12, 2026 AT 13:00OH MY GOD I JUST FOUND A PATIENT'S DIAGNOSIS LABELLED AS 'NORMAL' WHEN THE RADIOLOGIST CLEARLY WROTE 'SUSPECTED LUNG CANCER' IN THE NOTES. THIS IS A MURDER WAITING TO HAPPEN. WHO APPROVED THIS DATA? WHO IS GETTING PAID TO LET THIS SLIP THROUGH? I'M CALLING THE FDA RIGHT NOW.
Lauren Warner
January 12, 2026 AT 16:37Of course the labeling is garbage. You think a team of overworked temps with $15/hr contracts and zero medical training are going to get it right? This isn't a data problem. It's a moral failure. We're outsourcing life-or-death decisions to people who can't even spell 'hypertension'.
And now you want us to trust AI built on this? Please. The model doesn't know the difference between a tumor and a shadow because the people who labeled it didn't either.
beth cordell
January 13, 2026 AT 05:43YESSSS this is so real 😭 I work in a clinic and we had a model flag a patient for 'possible diabetes' but the label said 'no diabetes' because the annotator missed the HbA1c value in the note... we almost missed her diagnosis 😣
pls fix your labels ppl 🙏❤️
Craig Wright
January 15, 2026 AT 02:36It is regrettable that such a critical domain as medical data annotation is subjected to such amateurish practices. In the United Kingdom, we maintain rigorous standards for clinical documentation. The fact that boundary mislabeling reaches 41% in Australian datasets is not merely an oversight-it is a dereliction of professional duty.
One must ask: who is responsible for the accreditation of these annotators? What oversight bodies are being consulted? This is not a technical challenge-it is a failure of governance.
jordan shiyangeni
January 16, 2026 AT 03:40Let me be perfectly clear: the entire premise of using AI in medical diagnosis is fundamentally flawed if the training data is this sloppy. You're not building a model-you're building a liability. Every time a label is wrong, you're not just reducing accuracy-you're eroding the ethical foundation of medicine.
And don't give me that 'we're just a startup' excuse. This isn't about resources-it's about priorities. If you can't get the labels right, you shouldn't be touching patient data at all. The fact that you're even asking how to 'ask for corrections' instead of demanding perfection speaks volumes about your institutional incompetence.
MIT showed that fixing 5% of labels outperformed upgrading from ResNet-50 to ResNet-101. That's not a win. That's a confession that your entire AI pipeline was built on sand. And now you're gambling with lives because you didn't want to pay for proper annotation. You're not a data scientist. You're a negligence enabler.
Abner San Diego
January 17, 2026 AT 19:28Bro I just skimmed this and honestly I'm tired. We all know labels are trash. Everyone knows. But nobody does anything. The tools exist, the templates exist, the studies exist. So why are we still here? Why are we still arguing about whether a 4mm nodule is actually 8mm?
It's not the annotators. It's not the AI. It's the system. Nobody gets fired for bad labels. Nobody gets promoted for fixing them. So we keep pretending it's fine until someone dies. Then we move on to the next dataset.
Pass the popcorn.
Eileen Reilly
January 18, 2026 AT 13:33ok so i just used cleanlab on our radiology set and it flagged 142 examples as probably wrong but like half of them were just weird angles or artifacts?? i thought it was gonna be easy but now i have to go through every single one with a radiologist and its taking forever 😩
also why is everyone so mad? its just data lol
Monica Puglia
January 20, 2026 AT 06:15Hi everyone-just wanted to say thank you for sharing this. I work with a small team of volunteer annotators (mostly retired nurses and med students) and this guide has been a game changer 🙌
We started using the template you shared and now we have a 92% agreement rate between annotators. The key was adding real examples to the guidelines-not just definitions.
You don’t need a big budget. You just need to care. And you’re all caring. That’s everything.
❤️